
Location: DEI – Politecnico di Bari
Lab head: Prof. Vitoantonio Bevilacqua
Contact: vitoantonio.bevilacqua@poliba.it
Lab Team
Lab Mission
Setting up the INTelligent Optimized Computer-Aided Diagnosis Systems for bioengineers Laboratory (short INTOCADS) – AREA 4: Data and computational modelling. The laboratory will include a complete facility to design, implement and validate optimized decision support systems by means of the following workflow (materials and methods):
1) acquisition of biosignals and supply of bioimages and biodata for data-driven digital twins to simulate and estimate patient-specific characteristics of diseases;
2) advanced and intelligent techniques for processing and segmenting the available materials (biosignals,
bioimages, biodata and biomarkers) and forming the biopatterns;
3) intelligent and optimized methodologies for multifactorial and combined biopattern identification
(radiomic and radiogenomic feature extraction, reduction and transformation);
4) modelling, implementation, training and validation of machine/deep learning optimized topology for
providing Decision Support and Computer Aided Decision/Diagnosis Systems;
5) classification, characterization, staging and grading of pathologies (chronic, acute, neurodegenerative), or motor disorders.
Link: https://labinfind.poliba.it/ricerca-scientifica/pnrr/brief/intocads/
Scientific Publications
Zaccaria G.M., Altini, N., Mongelli, V., Marino, F., Bevilacqua, V. Development and validation of a machine learning prognostic model based on an epigenomic signature in patients with pancreatic ductal adenocarcinoma, 2025, p. 105883., doi: 10.1016/j.ijmedinf.2025.105883
Berloco, F., Zaccaria G.M., Altini, N., Colucci, S., Bevilacqua, V. A Multimodal Framework for Assessing the Link between Pathomics, Transcriptomics, and Pancreatic Cancer Mutations. Computerized Medical Imaging and Graphics, 2025, p. 102526. doi: 10.1016/j.compmedimag.2025.102526
Vladimiro Suglia, Cristian Camardella, Gianluca Rinaldi, Domenico Chiaradia, Domenico Buongiorno, Hui Zhou,Antonio Frisoli, Daniele Leonardis, Vitoantonio Bevilacqua. Muscle Network Analysis of a Dynamic Bilateral Task With an Upper Limb Exoskeleton- 19th International Conference on Rehabilitation Robotics (ICORR) May 12–16, 2025, Chicago, USA
Palazzo, L.; Suglia, V.; Grieco, S.; Buongiorno, D.; Brunetti, A.; Carnimeo, L.; Amitrano, F.; Coccia, A.; Pagano, G.; D’Addio, G.; Bevilacqua, V. A Deep Learning-Based Framework Oriented to Pathological Gait Recognition with Inertial Sensors. Sensors 2025, 25, 260. doi: 10.3390/s25010260
Zaccaria G.M., Berloco, F., Buongiorno, D., Brunetti, A., Altini, N., Bevilacqua, V. A time-dependent explainable radiomic analysis from the multi-omic cohort of CPTAC-Pancreatic Ductal Adenocarcinoma. Computer Methods and Programs in Biomedicine, 2024, p.108408. doi: 10.1016/j.cmpb.2024.108408
Zaccaria, G. M., Altini, N., Mezzolla, G., Vegliante, M. C., Stranieri, M., Pappagallo, S. A., Ciavarella, S., Guarini, A., Bevilacqua, V. SurvIAE: Survival prediction with Interpretable Autoencoders from Diffuse Large B-Cells Lymphoma gene expression data. Computer Methods and Programs in Biomedicine, 2024, 244, p.107966. doi: 10.1016/j.cmpb.2023.107966
Berloco, F., Marvulli, P.M., Suglia, V., Colucci, S., Pagano, G., Palazzo, L., Aliani., M., Castellana, G., Guido, P., D’Addio, G., Bevilacqua, V. Enhancing Survival Analysis Model Selection through XAI(t) in Healthcare. Applied Sciences, 2024, 14(14), 6084. doi: 10.3390/app14146084
Clemente, L., La Rocca, M., Paparella, G., Delussi, M., Tancredi, G., Ricci, K., Procida, G., Introna, A., Brunetti, A., Taurisano, P., Bevilacqua, V., de Tommaso, M. Exploring Aesthetic Perception in Impaired Aging: A Multimodal Brain—Computer Interface Study, 2024, Sensors, 24(7), 2329. doi: 10.3390/s24072329
Sibiliano, E., Buongiorno, D., Lassi, M., Grippo, A., Bessi, V., Sorbi, S., Mazzoni, A., Bevilacqua, V., Brunetti, A.Understanding the Role of Self-Attention in a Transformer Model for the Discrimination of SCD From MCI Using Resting-State EEG, 2024, 28(6):3422-3433. doi:110.1109/JBHI.2024.3390606
Suglia, V., Palazzo, L., Bevilacqua, V., Passantino, A., Pagano, G., D’Addio, G. A Novel Framework Based on Deep Learning Architecture for Continuous Human Activity Recognition with Inertial Sensors. Sensors, 2024, 24, 2199. doi: 10.3390/s24072199
Altini, N., Rossini, M., Turkevi-Nagy, S., Pesce, F., Pontrelli, P., Prencipe, B., Berloco, F., Seshan, S., Gibier, J. B., Dorado, A. P., Bueno, G., Peruzzi, L., Rossi, M., Eccher, A., Li, F., Koumpis, A., Beyan, O., Barratt, J., Huy Quoc, V., Mohan, C., Van Nguyen, H., Cicalese, P. A., Ernst, A., Gesualdo, L., Bevilacqua, V., Becker, J. U. Performance and Limitations of a Supervised Deep Learning Approach for the Histopathological Oxford Classification of Glomeruli with IgA Nephropathy. Computer Methods and Programs in Biomedicine, 2023, p.107814. doi: 10.1016/j.cmpb.2023.107814
Prencipe, B., Delprete, C., Garolla, E., Corallo, F., Gravina, M., Natalicchio, M. I., Buongiorno, D., Bevilacqua, V., Altini, N., Brunetti, A. An Explainable Radiogenomic Framework to Predict Mutational Status of KRAS and EGFR in Lung Adenocarcinoma Patients. Bioengineering. 2023; 10(7):747. doi: 10.3390/bioengineering10070747
Brunetti, A., Buongiorno, D., Altini, N., Bevilacqua, V. Enabling Technologies for Optimized Diagnosis, Therapy and Rehabilitation: Obtained Results and Future Works. In: Bochicchio, M., Siciliano, P., Monteriù, A., Bettelli, A., De Fano, D. (eds) Ambient Assisted Living. ForItAAL 2023. Lecture Notes in Bioengineering. Springer, Cham. doi: 10.1007/978-3-031-63913-5_19
Suglia, V., Brunetti, A., Pasquini, G., Caputo, M., Marvulli, T. M., Sibilano, E., Della Bella, S., Carrozza, P., Beni, C., Naso, D., Monaco, V., Cristella, G., Bevilacqua, V., Buongiorno, D. A Serious Game for the Assessment of Visuomotor Adaptation Capabilities during Locomotion Tasks Employing an Embodied Avatar in Virtual Reality.Sensors. 2023; 23(11):5017 doi: 10.3390/s23115017
Gentile, E., Brunetti, A., Ricci, K., Vecchio, E., Santoro, C., Sibilano, E., Bevilacqua, V., Iliceto, G., Craighero, L., de Tommaso, M. Effects of movement congruence on motor resonance in early Parkinson’s disease. Scientific Reports, 2023, 13(1), p.14887. doi: 10.1038/s41598-023-42112-2
Sibilano, E., Brunetti, A., Buongiorno, D., Lassi, M., Grippo, A., Bessi, V., Micera, S., Mazzoni, A., Bevilacqua, V. An attention-based deep learning approach for the classification of subjective cognitive decline and mild cognitive impairment using resting-state EEG. J Neural Eng. 2023 Feb 6. doi: 10.1088/1741-2552/acb96e
Devices

Computing Workstation for data analysis
This includes one workstation for high-performance computing, one workstation for networking functionalities and two mobile workstations to support research activities. All the workstations are equipped with high-end GPU.

Integrated system for high-density EEG/EMG and ECG signal acquisition and processing
This is based on the g.HIamp system, a versatile biosignal amplifier system with 256 channels for precise EEG, ECoG, ECG, EMG, and EOG measurements, both invasive and non-invasive. It ensures excellent signal resolution and wide sensitivity without saturation. With real-time signal processing and high-speed DSP, it offers fast and accurate data analysis. Compatible with various electrode types, it supports seamless integration with external body sensors. The system includes advanced artifact suppression features and can be easily configured for different channel counts and electrode setups, making it ideal for a wide range of research and clinical applications.
Data sheet: download

Motion Tracking System
This includes a system for the analysis of human movement, for diagnostic and therapeutic purposes, both from a kinematic point of view (with and without markers) and from a kinetic point of view complete with:
- Vicon optical and marker-based motion analysis system complete with 8 Vero2.2 cameras with tripods, two full HD RGB cameras, accessories, Mocap suits and software licenses for data acquisition management.
Data sheet: download
- Two portable 3D dynamometer platforms AMTI mod. Accugait and software license for managing kinematic data acquisition.
Data sheet: download
- Xsens inertial non-marker based motion analysis system complete with 18 MTw Awinda Wireless 3DOF Motion Tracker, Manus VR Sensored Gloves – Xsens Pro Metagloves and a license MVN Analyze Pro Lifetime for managing data acquisition and processing.
Data sheet: download
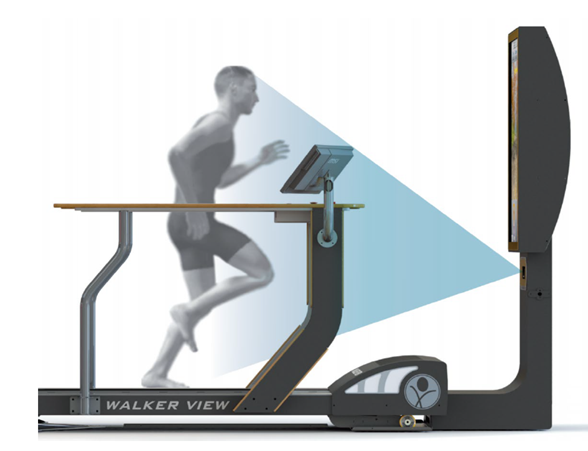
Walker View MED 4.0 SCX System
This is a system for the postural analysis of Run Analysis and Gait Analysis through the generation of immediate and objective visual feedback. Walker View MED 4.0 SCX, developed by TecnoBody research, analyses athletic performance and walking gait comprehensively, providing postural analysis reports for rehabilitation and sports medicine. With immediate feedback, it enhances posture, lower limb loading, and more during both walking and running. The SCX Speed Control feature revolutionizes treadmill training by adapting to users’ pace and stride, ushering in a new era in gait rehabilitation and running.
Data sheet: download

Hololens
Microsoft HoloLens 2, created and produced by Microsoft, is a mixed reality head-mounted display. HoloLens 2 utilizes a combination of waveguide and laser-based technology to deliver stereoscopic and full-colour mixed reality experiences.
Data sheet: link











